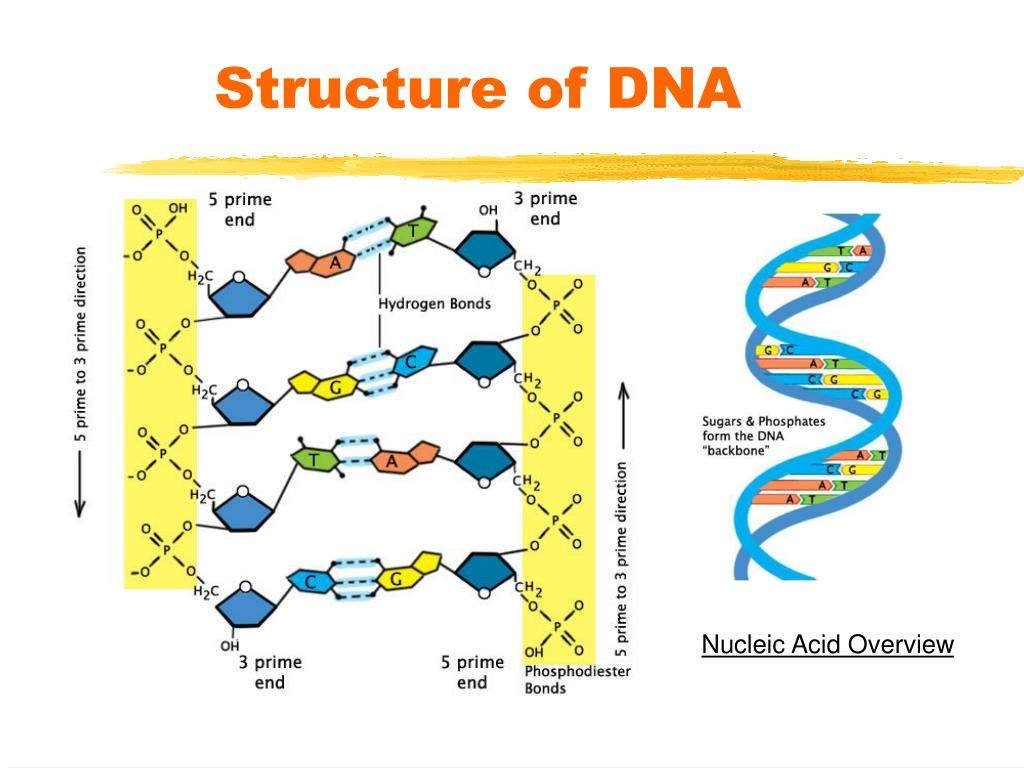
How to make dancing dna
Video of ‘dancing DNA’ developed by researchers | UCL News
Videos showing for the first time how small circles of DNA adopt dance-like movements have been developed by a team led by researchers at UCL and the Universities of Leeds, York and Sheffield.
The footage is based on some of the highest resolution images of a single molecule of DNA ever captured, with DNA seen to “dance” in microscopy data recorded at the London Centre for Nanotechnology at UCL.
The images show in unprecedented detail how the stresses and strains that are placed on DNA when it is crammed inside cells can change its shape.
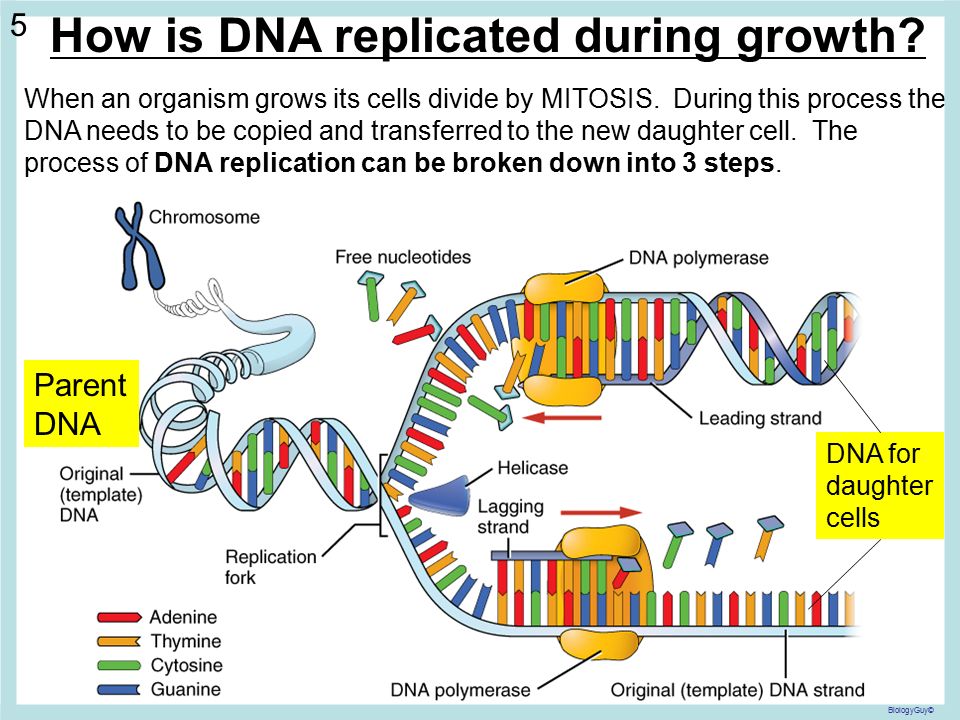
Previously scientists were only able to see DNA by using microscopes limited to taking static images. In the new study, published in Nature Communications, the research team combined advanced atomic force microscopy pioneered with supercomputer simulations to create videos of twisted molecules of DNA.
Co-lead author Dr Alice Pyne (University of Sheffield and the London Centre for Nanotechnology at UCL) said: "Seeing is believing, but with something as small as DNA, seeing the helical structure of the entire DNA molecule was extremely challenging.
"The videos we have developed enable us to observe DNA twisting in a level of detail that has never been seen before."
Co-author Professor Bart Hoogenboom (London Centre for Nanotechnology at UCL) said: "These high-resolution images of DNA are a culmination of many years of microscopy research at the London Centre for Nanotechnology. The great advance here is that we have been able to match them with computational modelling by our collaborators, thus enabling us to provide an interpretation of how DNA twists down to the atomic details."
The images are so detailed it is possible to see the iconic double helical structure of DNA, but when combined with the simulations, the researchers were able to see the position of every single atom in the DNA and how it twists and writhes.
Dr Agnes Noy, of the University of York, who did the theoretical modelling in the study, said: “The computer simulations and microscopy images agree so well that they boost the resolution of experiments and enable us to track how each atom of the double helix of DNA dances.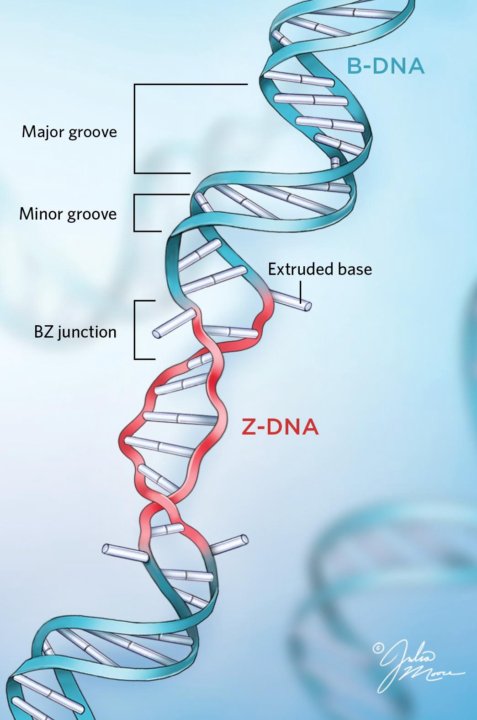 "
"
Every human cell contains two metres of DNA. In order for this DNA to fit inside our cells, it has evolved to twist, turn and coil – a process called supercoiling. That means that loopy DNA is everywhere in the genome, forming twisted structures which show more dynamic behaviour than their relaxed counterparts.
To investigate how this process works, the research team studied small "packets" of genetic information called DNA minicircles, engineered and isolated from bacteria. DNA minicircles are special because the molecule is joined at both ends to form a loop. This loop enabled the researchers to give the DNA minicircles an extra added twist, making the DNA "dance" more vigorously.
When the researchers imaged relaxed DNA, without any twists, they saw that it did very little. But when they gave the DNA an added twist, it suddenly became far more dynamic and could be seen to adopt some very exotic shapes.
These exotic "dance moves" were found to be the key to finding binding partners for the DNA.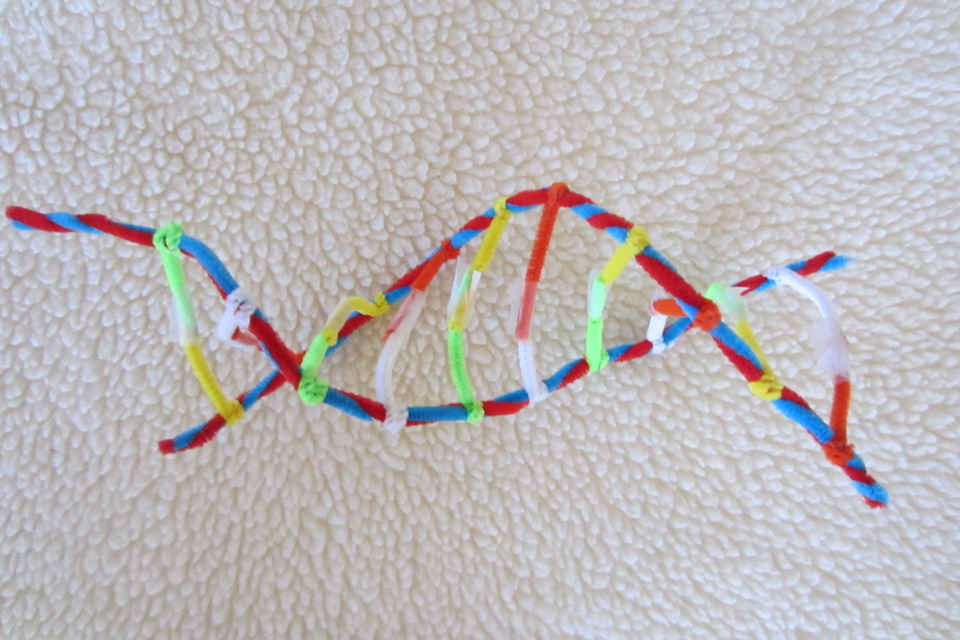
Gene therapy is the use of nucleic acids such as DNA to repair, replace, or regulate genes to prevent or treat human disease. In the past few decades, hundreds of gene therapy candidate genes have been uncovered, yet very few of these have turned into target therapies because of the challenge of delivering the gene therapy.
Dr Sarah Harris, of the University of Leeds, who supervised the research, said: "The laws of physics apply just as well to the molecules that make up living systems as to sub-atomic particles and galaxies. The synergy between our experiments and computer models shows we are beginning to understand the physics underlying supercoiled DNA.”
The study also involved researchers from the John Innes Centre, Norwich, the University of Liverpool, and the Baylor College of Medicine in Houston, Texas.
Links
- Full paper in Nature Communications
- Professor Bart Hoogenboom’s academic profile
- London Centre for Nanotechnology
- UCL Physics & Astronomy
- UCL Mathematical & Physical Sciences
Image
Credit: University of Sheffield for image and video.
Media contact
Mark Greaves
T: +44 (0)7990 675947
E: m.greaves [at] ucl.ac.uk
Visualisation of 'dancing DNA' | University of Leeds
- Category
- Science news
- Date
Videos showing for the first time how small circles of DNA adopt dance-like movements inside a cell have been developed by researchers in Yorkshire.
The footage, created by a team of scientists from the Universities of Leeds, Sheffield and York, and recorded at UCL, is based on the highest resolution images of a single molecule of DNA ever captured.
They show in unprecedented detail how the stresses and strains that are placed on DNA when it is crammed inside cells can change its shape.
Previously scientists were only able to see DNA by using microscopes limited to taking static images. But now the Yorkshire team has combined advanced atomic force microscopy with supercomputer simulations to create videos of twisted molecules of DNA.
The images are so detailed it is possible to see the iconic double helical structure of DNA, but when combined with the simulations, the researchers were able to see the position of every single atom in the DNA and how it twists and writhes.
Credit: University of Sheffield
Dr Alice Pyne, Lecturer in Polymers & Soft Matter at the University of Sheffield, who captured the footage, said: "Seeing is believing, but with something as small as DNA, seeing the helical structure of the entire DNA molecule was extremely challenging.
"The videos we have developed enable us to observe DNA twisting in a level of detail that has never been seen before."
The study, Base-pair resolution analysis of the effect of supercoiling on DNA flexibility and major groove recognition by triplex-forming oligonucleotides, is published in Nature Communications.
Every human cell contains two metres of DNA. In order for this DNA to fit inside our cells, it has evolved to twist, turn and coil a process called supercoiling. That means that loopy DNA is everywhere in the genome, forming twisted structures which show more dynamic behaviour than their relaxed counterparts.
To investigate how this process works, the research team studied small "packets" of genetic information called DNA minicircles, engineered and isolated from bacteria. DNA minicircles are special because the molecule is joined at both ends to form a loop. This loop enabled the researchers to give the DNA minicircles an extra added twist, making the DNA "dance" more vigorously.
When the researchers imaged relaxed DNA, without any twists, they saw that it did very little. But when they gave the DNA an added twist, it suddenly became far more dynamic and could be seen to adopt some very exotic shapes.
These exotic "dance moves" were found to be the key to finding binding partners for the DNA.
How the findings aid future research
Gene therapy is the use of nucleic acids such as DNA to repair, replace, or regulate genes to prevent or treat human disease. In the past few decades, hundreds of gene therapy candidate genes have been uncovered, yet very few of these have turned into target therapies because of the challenge of delivering the gene therapy.
Professor Lynn Zechiedrich from Baylor College of Medicine in Houston Texas, who made the minicircles used in this study, has found a way to design supercoiled minicircles or, minivectors for use in gene therapy by inserting short genetic messages.
Professor Zechiedrich said: "The research team in Yorkshire have developed a technique that reveals in remarkable detail how wrinkled, bubbled, kinked, denatured, and strangely shaped they are!
"We have to understand how supercoiling, which is so important for DNA activities in cells, affects DNA in hope that we can learn how to mimic or control it someday.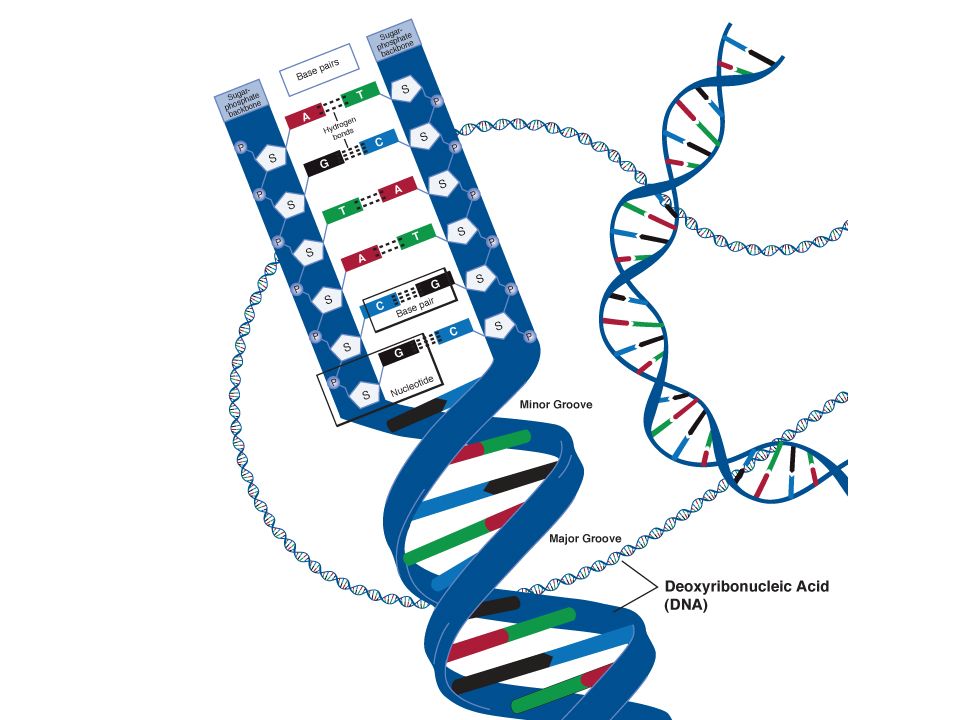 "
"
Dr Massa Shoura from Stanford University, who has detected DNA minicircles in human cells, said: "Very little is currently understood about the function of circular DNAs in cells, but there is a chance they could be used as markers for early detection of disease."
Dr Sarah Harris, Associate Professor in the School of Physics and Astronomy at the University of Leeds, who supervised the research, said: "The laws of physics apply just as well to the molecules that make up living systems as to sub-atomic particles and galaxies. The synergy between our experiments and computer models shows we are beginning to understand the physics underlying supercoiled DNA. This insight should help researchers such as Professor Zechiedrich design bespoke minicircles for future therapies."
Further information
- Top image: A visualisation of a DNA minicircle
- For media enquiries, please contact David Lewis in the University of Leeds press office: d.lewis@leeds.
 ac.uk
ac.uk
Nobel Prize in Chemistry - from award to award
In the time since the current Nobel Prize winners in Chemistry were announced, biologists have found a new DNA repair enzyme, learned how proteins look for mismatches in DNA, and seen how damaged DNA moves in human cells.
On October 6, we learned about who this year won the Nobel Prize in Chemistry, and hastened to inform our readers about it. Recall that the prize was awarded for research in the field of DNA repair. nine0003
Base excision repair – only one corrupted genetic “letter” is excised pointwise from DNA. Thomas Lindahl received the Nobel Prize for deciphering the mechanism of such reparation. (Illustration by the Nobel Committee.)
Nucleotide excision repair, "according to Sanjar" - a whole piece with a damaged "letter" is cut out of DNA. (Illustration by the Nobel Committee.)
Mismatched nucleotide repair is triggered by errors in the DNA copying machine, the mechanism of which was deciphered by Paul Modrich - special proteins detect a fragment with mismatched nucleotides and remove it so that a 9 appears in its place0003
Schematic of the structure of a nucleotide - a three-part molecule, which may include various nitrogenous bases that act as genetic "letters". (Illustrated by Wikipedia.)
(Illustrated by Wikipedia.)
A new glycosylase (left), which removes modified bases in the base excision repair mechanism, does not turn them out of the double strand like other glycosylases (right), but leaves it inside the DNA helix. (Illustration Brandt Eichman / Van
Between 80% and 90% of cancer cases are due to malfunctions in DNA repair systems. (Photo: Prostate cancer cell.) (Photo by Visuals Unlimited / Corbis.)
‹
›
View full size
In our heredity molecule, defects often occur that can have the most unpleasant effect on the state of the body: DNA can simply break in one or two chains at once, and then it just needs to be sewn back, but there are also more ingenious damage associated with the modification of the letters of the genetic code - nucleotides, or with the replacement of correct nucleotides with incorrect ones, distorting the meaning of the information recorded in DNA. Here, to correct such modifications and inappropriate replacements, there are special enzyme systems, and it is for deciphering how they work that Thomas Lindahl, Aziz Sanjar and Paul Modric will receive their Nobel Prize on December 10th. nine0003
nine0003
Two months from the announcement of the laureates to the awards ceremony, on the one hand, is a short period, but only if you do not know what new things we have learned about DNA repair during this time. And we learned quite a lot. For example, that human DNA in the region of double-stranded breaks begins to actively twitch, wriggle, in general, its mobility noticeably increases, as if the molecule deliberately swings its suddenly formed ends. Until recently, such "dancing DNA" was observed in various other cells, and now they finally saw the same thing in human ones. nine0003
Moreover, the team from the Rockefeller Institute, who published their discovery in Cell , at the same time identified those intracellular structures that increase the mobility of DNA - they turned out to be protein complexes of the nuclear envelope and the microtubules of the cytoplasm connected to them. (Microtubules are long rod-shaped structures made up of many protein molecules and playing the role of a cell skeleton. ) That is, DNA that has breaks in both chains begins to flicker under the action of a complex protein apparatus, and this is necessary for the repair to be successful - it is believed that if the DNA at the break site is actively moving, then it will be easier for repair enzymes to find and connect the broken ends. nine0003
) That is, DNA that has breaks in both chains begins to flicker under the action of a complex protein apparatus, and this is necessary for the repair to be successful - it is believed that if the DNA at the break site is actively moving, then it will be easier for repair enzymes to find and connect the broken ends. nine0003
As for the very mechanism of stitching gaps, biologists continue to clarify. For example, in an article in Nature Cell Biology , researchers at the University of Alberta describe the behavior of the RNF138 protein, whose job is to remove another protein, Ku, from the resulting DNA ends. Ku serves as a kind of plug for them, but in his presence it is impossible to close the gap, which is why RNF138 is needed, about which not much is known yet.
If we talk about the actual "Nobel" mechanisms of DNA repair, then here it is worth recalling the work of biologists from Vanderbilt University, who discovered another protein involved in the so-called excision repair by removing damaged bases. When a modified nucleotide appears in DNA, glycosylase enzymes come into play, which cleave off the damaged nitrogenous base from the nucleotide (recall, just in case, that a nucleotide is a three-part molecule consisting of a phosphoric acid residue, a deoxyribose sugar and one of the four nitrogenous bases; in the composition DNA includes 4 bases, adenine, thymine, guanine and cytosine, and they are the very 4 letters of the genetic code). nine0003
When a modified nucleotide appears in DNA, glycosylase enzymes come into play, which cleave off the damaged nitrogenous base from the nucleotide (recall, just in case, that a nucleotide is a three-part molecule consisting of a phosphoric acid residue, a deoxyribose sugar and one of the four nitrogenous bases; in the composition DNA includes 4 bases, adenine, thymine, guanine and cytosine, and they are the very 4 letters of the genetic code). nine0003
The mechanism of base excision repair eliminates a whole class of damage, and therefore glycosylases (the first of which were described by the current laureate Thomas Lindahl) exist not one or two. However, the enzyme, which is described in Nature , is different from all its other "colleagues". Normally, glycosylases find a damaged base, rip it off its partner in the other strand, and twist it out of the double helix, and that protruding base is chemically cleaved from the DNA. The new glycosylase, called AlkD, eliminates alkylated bases (that is, those to which a hydrocarbon chemical group is attached), but it does not turn the modified base outward (see figure), and the damaged area itself does not recognize directly, but by indirect signs, and has dealing with more bulky modifications than other related enzymes. nine0003
nine0003
In order to repair any damage in DNA, you must first find it. And this is not an easy task, given the millions and billions of "letters" that make up the genomes of living beings. Obviously, DNA repair proteins must have some special search strategies - for example, the repair system can more carefully monitor the part of the genome where the likelihood of mutations is highest. The easiest way to implement such a strategy is for mismatch repair enzymes, the mechanism of which was studied by Paul Modric. nine0003
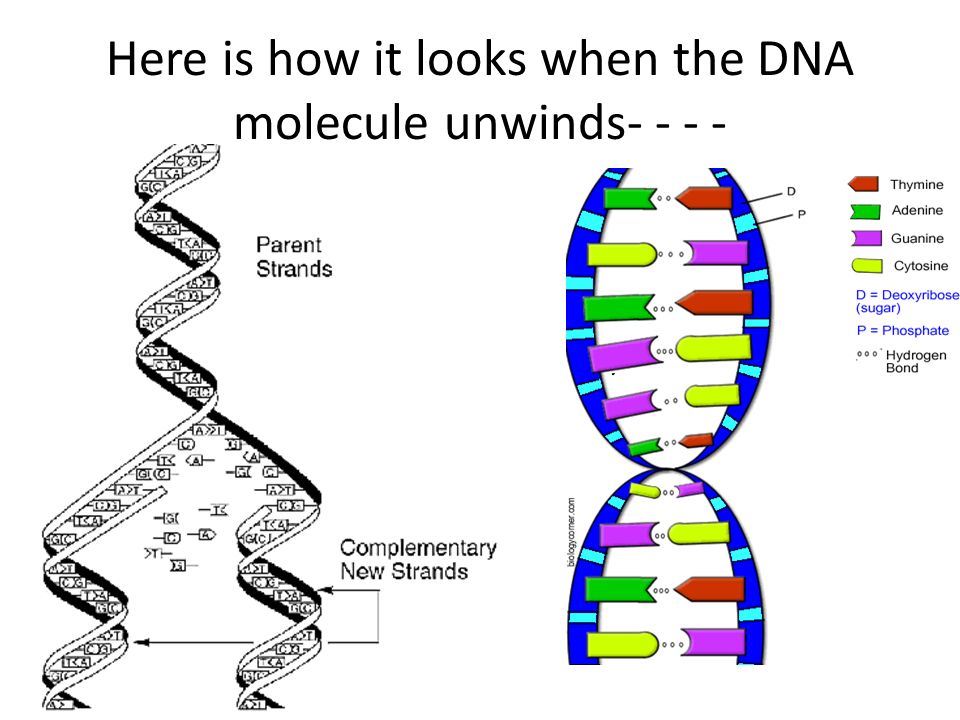
Mismatch repair corrects the errors left behind by the replication machinery, i.e. DNA doubling. As you know, the genetic "letters"-bases are located opposite each other in two DNA strands, not by chance, but according to the rule of complementarity: adenine is opposite to thymine, guanine is opposite to cytosine. When DNA is duplicated, the chains of the old molecule are separated, and on each of them a new - daughter - chain is synthesized, so that its nucleotides are complementary to the nucleotides of the old, maternal chain. But it happens that the enzymes synthesizing new chains make mistakes, and put in the new chain the wrong “letter” that is needed there - a mismatch happens. This happens relatively rarely - once every 30-60 million bases - but a mutation is a mutation, and it needs to be corrected. nine0003
But it happens that the enzymes synthesizing new chains make mistakes, and put in the new chain the wrong “letter” that is needed there - a mismatch happens. This happens relatively rarely - once every 30-60 million bases - but a mutation is a mutation, and it needs to be corrected. nine0003
The recognition of mismatched nucleotides is handled by the MutS protein, and, as the experiments of the research group from the University of Michigan showed, MutS does not patiently scan all the bases in DNA at all, but quickly skips most of them until it reaches the place where replication actually occurs, and here its progress is greatly slowed down - the protein begins to look for bad nucleotide pairs. The strategy, in general, is obvious, but experimental confirmation that this is exactly what is happening has only now been obtained. Experiments described in Proceedings of the National Academy of Sciences , was carried out on bacteria, but the authors of the work believe that in humans, the search for incorrect nucleotide pairs proceeds in exactly the same way - the mismatch repair system in the living world is quite conservative.
Proteins that repair DNA do not exist on their own, they have many "helpers" in the cell, and these "helpers" are still not known to everyone. For example, researchers at the University of Copenhagen recently published in Nature article discussing how h2 histone helps DNA repair. In fact, if not histones, special packaging proteins that are constantly associated with cellular DNA and determine the spatial structure and activity of certain sections of it, who can not be helpers here, if not histones. Histones should be among the first to feel damage in it. Indeed, when many breaks appear in DNA, they send an alarm signal, but until recently nothing was known about a specific h2 protein. nine0003
It is also worth mentioning to whom the histones report information about the poor state of the genetic molecule: among their “correspondents” is the legendary p53 protein, the “guardian of the genome,” as it is sometimes called. When damage occurs in the DNA, it inhibits division and forces the repair systems to fix whatever is needed; if there is a lot of damage, it launches a program of apoptosis, cell suicide, so that the cell, which is good, does not turn into a cancer cell due to mutations. When something is wrong with p53 itself, the risk of developing a tumor is high, and indeed, a lot of cancers arise due to malfunction or absence of p53. nine0003
When something is wrong with p53 itself, the risk of developing a tumor is high, and indeed, a lot of cancers arise due to malfunction or absence of p53. nine0003
It is not yet clear how cancer cells themselves manage to cope with DNA damage, but we are gradually learning more about it: in the journal Cancer Cell , specialists from the Massachusetts Institute of Technology describe a rather complex scheme of interaction of several cellular proteins that allow tumor cells to survive in the absence of p53. The bottom line is that they start a special system that allows you to slow down the division and repair such damage in DNA that even a tumor could not survive. nine0003
In general, it is difficult to talk about DNA repair without mentioning cancer - it is believed that from 80 to 90% of oncological diseases happen just because of the incorrect operation of repair machines, and a lot of work is published on the relationship of one to the other. Naturally, everyone is trying to figure out how to make a drug that would act on cancer through repair mechanisms; the situation is complicated by the fact that repair systems often play in favor of the tumor.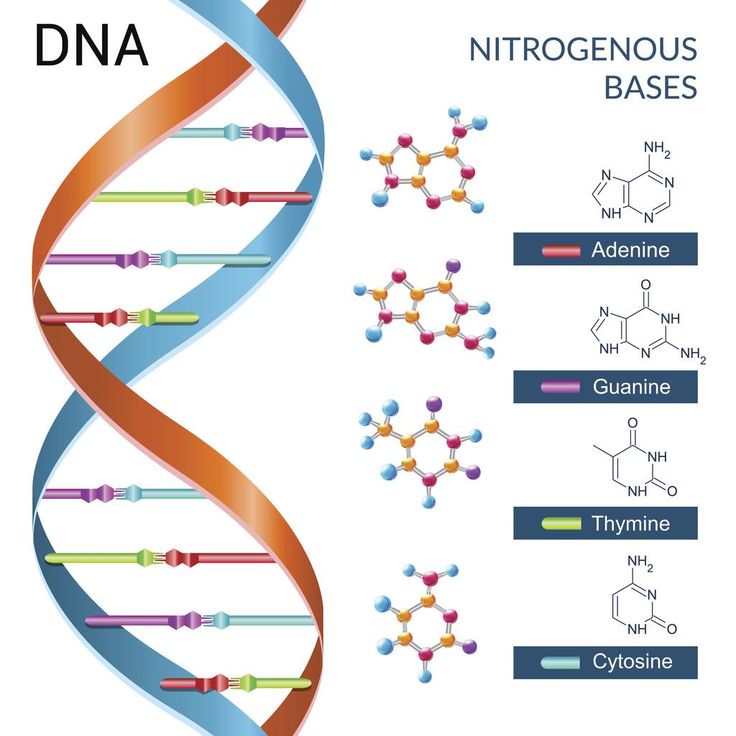 Many chemotherapy drugs are designed to destroy the DNA of cancer cells, and repair enzymes just do not allow this, so in order to successfully fight cancer, sometimes it is necessary to somehow suppress DNA-repairing genes and proteins in malignant cells. But, in order not to completely confuse readers with a huge mass of information, we allow ourselves to remain silent about such studies. nine0061
Many chemotherapy drugs are designed to destroy the DNA of cancer cells, and repair enzymes just do not allow this, so in order to successfully fight cancer, sometimes it is necessary to somehow suppress DNA-repairing genes and proteins in malignant cells. But, in order not to completely confuse readers with a huge mass of information, we allow ourselves to remain silent about such studies. nine0061
Non-canonical DNA
Article for the Bio/Mol/Text competition: We invite the reader to find out how thymine differs from uracil. Trivial answers - for example, " methyl group " or " in RNA uracil, and in DNA - thymine " are not accepted. In search of an answer, one will have to delve into the field of epigenetics in order to first understand the differences between cytosine and 5-methylcytosine. We'll have to learn about ultrasound, which can break DNA in a non-random way. To meet a fantastic tautomer that can be beneficial, but continues to harm. To see clear signs of tautomers in the NMR spectrum, until recently considered something indecent. We will also talk about the vandals' plans to destroy the foundation of epigenetics. You will be asked to fight the vandals or join them (both options are more interesting than standing on the sidelines). In conclusion, the authors reassure readers and apologize for the unintentional misleading. nine0003
To see clear signs of tautomers in the NMR spectrum, until recently considered something indecent. We will also talk about the vandals' plans to destroy the foundation of epigenetics. You will be asked to fight the vandals or join them (both options are more interesting than standing on the sidelines). In conclusion, the authors reassure readers and apologize for the unintentional misleading. nine0003
This work won first place in the Own Work nomination of the Bio/Mol/Text contest -2020/2021.
The partner of the nomination is the Russian Science Foundation.
The general partner of the competition is the annual biotechnology conference BiotechClub , organized by the international innovative biotechnology company BIOCAD .
Contest Sponsored by SkyGen: Leading Product Distributor for life science in the Russian market.
The sponsor of the competition is the Diaem company: the largest supplier of equipment, reagents and consumables for biological research and production.
"Book" sponsor of the competition - "Alpina non-fiction"
Chemistry is the science of substances; and since molecular biology studies substances and does not trade them as commodities, it needs a good knowledge of chemistry.
Erwin Chargaff. Beliberdin pandemonium
What's the difference?
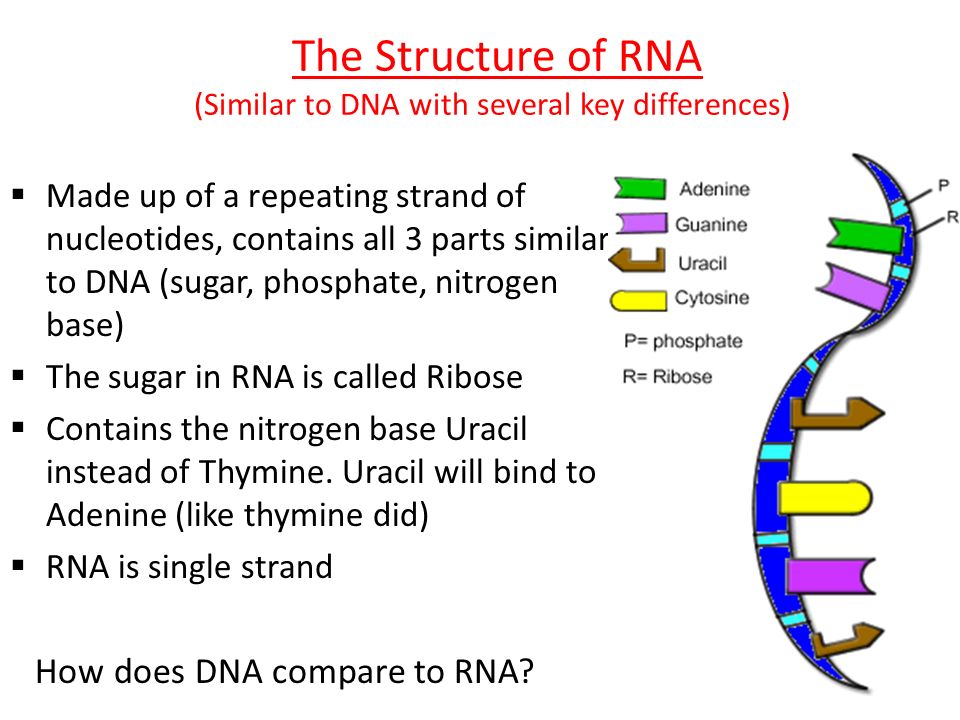
Since we wrote this text specifically for "Biomolecules" , we will not be surprised if you, dear reader, have heard, read or even taken an exam question about how DNA differs from RNA. We won’t be surprised if you not only passed the exam, but also have to take these exams yourself, prepare for lectures, reread textbooks. You probably know that thymine is in DNA, but uracil is in RNA. What is the difference between thymine and uracil? Of course, you remember that thymine has a methyl group. It would be more accurate to say that thymine is 5-methyluracil. But this is an incomplete answer to the question. We should be interested in what exactly does this methyl group in uracil change, how is it useful for DNA? This question could be answered in the language of chemistry: if the properties of DNA changed from replacing thymine with uracil, then these differences would need to be studied.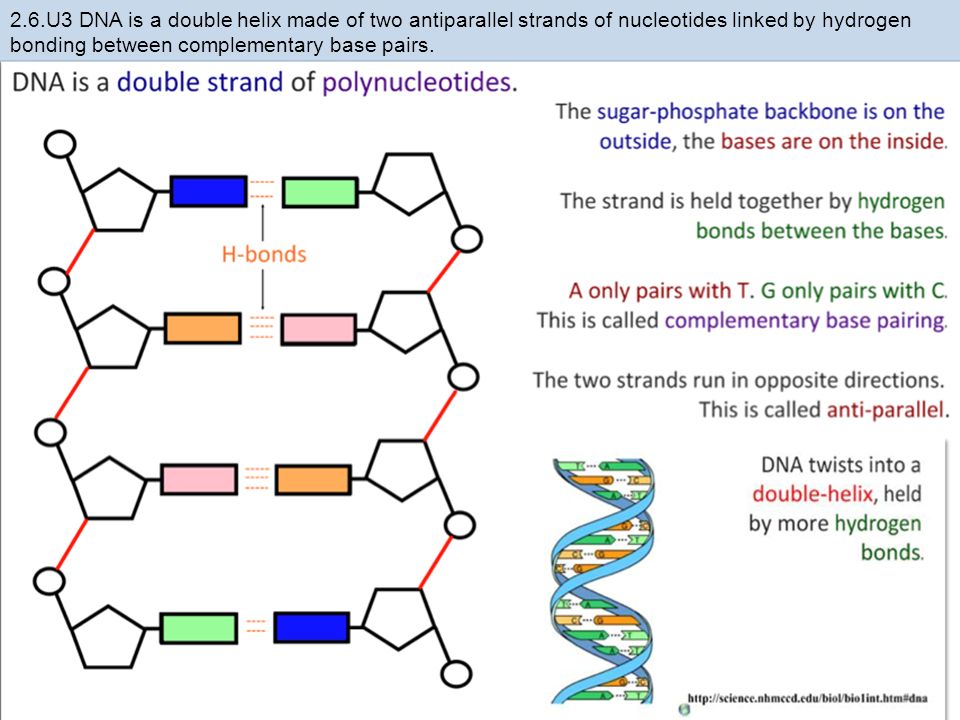 It is necessary and possible - it does not mean done yet. Answer to question " What is the difference between thymine and uracil? " has not been fully disclosed in the most popular molecular biology textbooks. If you read our text to the end, then we promise to answer this question. And you can surprise the examiner. Or you can surprise students at your lecture, which is perhaps even more useful.
It is necessary and possible - it does not mean done yet. Answer to question " What is the difference between thymine and uracil? " has not been fully disclosed in the most popular molecular biology textbooks. If you read our text to the end, then we promise to answer this question. And you can surprise the examiner. Or you can surprise students at your lecture, which is perhaps even more useful.
If we know how the methyl group in 5-methyluracil (that is, thymine) affects the properties of DNA, does this change our view? What is the difference? The main difference is in the ability to find the cause of the changes and evaluate the benefits of them. If we look at methyl groups in DNA as a stucco decoration on the wall of the Bolshoi Theater hall, then two fundamentally different points of view are possible. The first calms our curiosity by asserting that stucco decoration is needed simply “for beauty”. And the second point of view wins if you set up an experiment and remove the decor, replacing it with a bas-relief with portraits of Lenin and Stalin. It turns out that the echo in the concert hall will be reflected from the high foreheads of the Soviet leaders and spoil the acoustics of the hall. And suddenly there is an understanding that beauty was not accidental and carried great practical benefits. But in order to understand the benefits and for a full perception of beauty, the organizers of the repair of the Bolshoi Theater in 19The 1930s lacked primary education. The lack of education was fully compensated by the ability to admit their mistakes and learn from them. The stucco decoration was restored from photographs.
It turns out that the echo in the concert hall will be reflected from the high foreheads of the Soviet leaders and spoil the acoustics of the hall. And suddenly there is an understanding that beauty was not accidental and carried great practical benefits. But in order to understand the benefits and for a full perception of beauty, the organizers of the repair of the Bolshoi Theater in 19The 1930s lacked primary education. The lack of education was fully compensated by the ability to admit their mistakes and learn from them. The stucco decoration was restored from photographs.
Unfortunately, in molecular biology one can still find such a contemplative attitude towards certain chemical modifications. Does the example with thymine and uracil not seem as funny to you as the example with the bas-reliefs of leaders in the Bolshoi Theater? Then: Welcome to epigenetics ! nine0003
Encouraged by the opportunity to immediately learn something interesting and important, epigeneticists skip over the boring details of the influence of the methyl group on the chemistry of cytosine. Why know how an engine works when you're driving a new car that takes you down a new road straight into the castles in the air of a new and trendy science? In order not to go far, let's turn to the articles on epigenetics on "Biomolecule" . Epigenetics builds bridges from genetics to psychology [1], [2], studies diseases [3], [4] and promises to defeat aging [5–7]. Two of the seven articles even have chemical formulas [3], [4]! We recommend reading them to capture the beauty of contemplating the DNA methyl decor. nine0003
Why know how an engine works when you're driving a new car that takes you down a new road straight into the castles in the air of a new and trendy science? In order not to go far, let's turn to the articles on epigenetics on "Biomolecule" . Epigenetics builds bridges from genetics to psychology [1], [2], studies diseases [3], [4] and promises to defeat aging [5–7]. Two of the seven articles even have chemical formulas [3], [4]! We recommend reading them to capture the beauty of contemplating the DNA methyl decor. nine0003
What is the difference between cytosine and 5-methylcytosine?
Putting questions instead of headings in sections is ugly. We recognize this and repent. In our defense, we want to say that we tried to draw attention to the similarity of the problem of comparing cytosine and 5-methylcytosine on the one hand and the problem of comparing uracil and thymine (5-methyluracil). In order to make the similarity obvious, we reflected it in Figure 1. We are aware that each formula placed in the text of a popular science article reduces the number of readers by half. Four chemical formulas in one picture should deprive us of a readership. Not without reason, in articles on epigenetics on "Biomolecule" formulas are so rare. However, we ask you to make an effort and continue reading after looking at this drawing.
We are aware that each formula placed in the text of a popular science article reduces the number of readers by half. Four chemical formulas in one picture should deprive us of a readership. Not without reason, in articles on epigenetics on "Biomolecule" formulas are so rare. However, we ask you to make an effort and continue reading after looking at this drawing.
Figure 1. Uracil and thymine (top), cytosine and 5-methylcytosine (bottom). Spot the differences.
Wikipedia
Remember that you get the opportunity to surprise the examiner. Even two examiners: the first in molecular biology, and the second in epigenetics. If you have not yet chosen a special course in epigenetics, then we advise you to choose. If your university does not yet have such a special course, then you can surprise teachers of online courses with your knowledge. We are agitating everyone: come to this field of science, because, in our opinion, it is on the verge of amazing events! nine0003
At present, the seemingly impossible is combined in epigenetics: the molecular nature of the studied object is combined with naive descriptiveness and even some kind of childish belief in the purposefulness of all changes observed in DNA.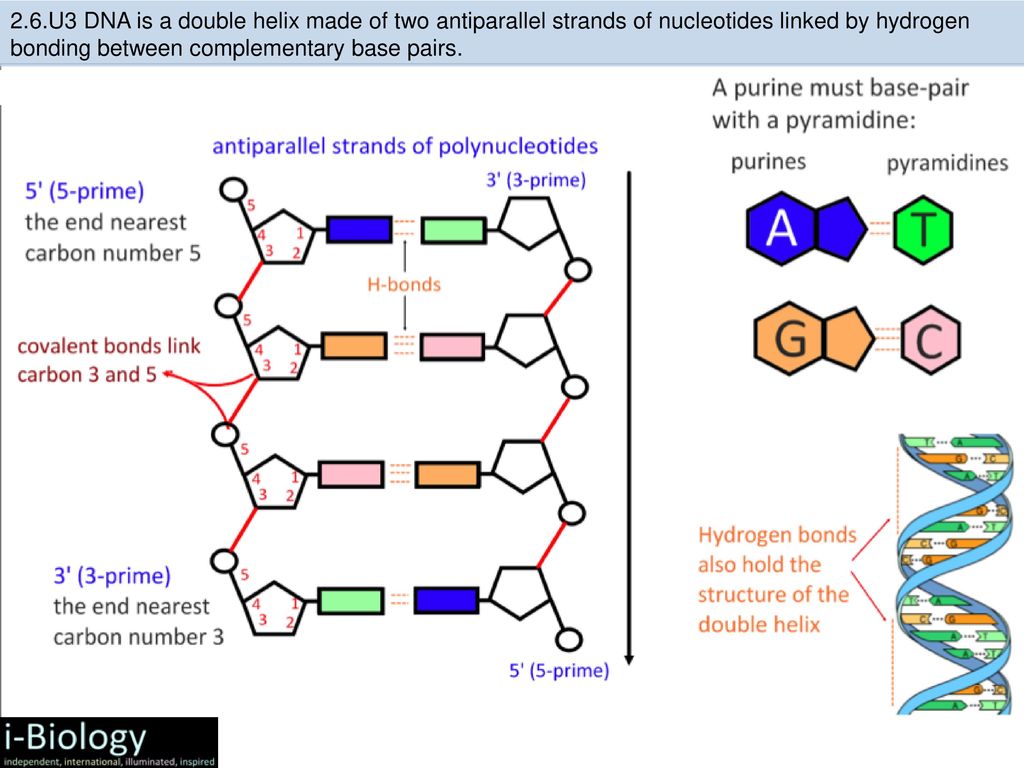 We changed the composition of the feed for mice with a predisposition to diabetes - and saw methyl groups in DNA. We trained lazybones for half a year - and also saw a change in the methylation of almost a thousand genes. The scope and scale of costs is almost inversely proportional to the predictive and explanatory capabilities of the work. nine0003
We changed the composition of the feed for mice with a predisposition to diabetes - and saw methyl groups in DNA. We trained lazybones for half a year - and also saw a change in the methylation of almost a thousand genes. The scope and scale of costs is almost inversely proportional to the predictive and explanatory capabilities of the work. nine0003
Unlike the methyl group in thymine, the methyl group of 5-methylcytosine is associated with important biological effects: a methyl group appears in the regulatory region of DNA and gene reading stops. Or here's another example: if a lot of 5-methylcytosines are found in DNA, then this is a sign of cancer. It would seem that this is a reason to study what changes in DNA when a methyl group appears. If you look into a good textbook of molecular biology (for example, Alberts' Molecular Biology of the Cell, 2013 edition, section 7.1), you will see that it is generally accepted that nothing in DNA changes [8]. More precisely, the authors of the textbook consistently develop the idea that the methyl group is enough for some proteins to recognize the desired site, while others do not.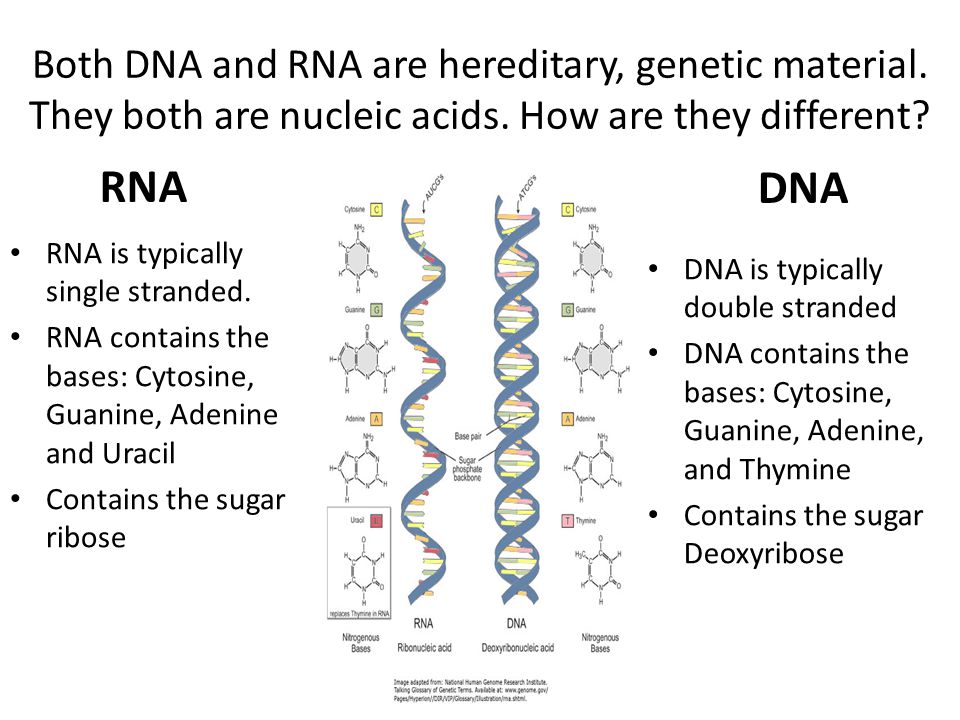 In mathematical statistics, this is called the "null hypothesis": in the first approximation, it is assumed that there are no changes and differences. Everyone who was forced to statistically process the results of their own experiments tested the null hypothesis. If you remember, all the fun begins if the null hypothesis can be rejected. nine0003
In mathematical statistics, this is called the "null hypothesis": in the first approximation, it is assumed that there are no changes and differences. Everyone who was forced to statistically process the results of their own experiments tested the null hypothesis. If you remember, all the fun begins if the null hypothesis can be rejected. nine0003
Why might methylation change nothing in DNA? Well, for example, because the methyl group is small, uncharged, it does not form hydrogen bonds. If methylation suddenly changes the properties of DNA in the same place, then this opens up completely different perspectives: at least the prospect of rewriting section 7.1 in the mentioned textbook.
It turns out he does! After methylated cytosines, DNA is 1.5 times more likely to be broken by ultrasound and other mechanochemical methods [9–13] (Fig. 2). nine0003
DNA properties change locally. The null hypothesis can be rejected and replaced with a more interesting picture! It's great, it's supposed to mean something, but what? In order to understand what happens in DNA as a result of cytosine methylation, it is necessary to explain what we know about the destruction of DNA by ultrasound. It turns out that DNA always breaks better after cytosine (even before methylation), and the easiest way for a break to form is between two adjacent “letters” CG (Fig. 2). The most perceptive exclaimed now: " So it is methylated there! ", and they will be right! It is there that cytosine is methylated, and DNA begins to break another 1.5 times more often.
It turns out that DNA always breaks better after cytosine (even before methylation), and the easiest way for a break to form is between two adjacent “letters” CG (Fig. 2). The most perceptive exclaimed now: " So it is methylated there! ", and they will be right! It is there that cytosine is methylated, and DNA begins to break another 1.5 times more often.
Figure 2. Relative frequency of DNA single-strand breaks between two adjacent nucleotides. CpG leads even without methylation!
Surprisingly, we were lucky precisely because when mechanochemical DNA cleavage was introduced as a way to prepare a sample for NGS sequencing, no one tried to test the “null hypothesis”. For some reason, the developers thought that DNA should be torn randomly. This seemed obvious, because DNA breaks along the sugar-phosphate backbone; What is the sequence of nucleotides here? This approach is called the "economy of thought" and is applicable to almost everything. Who would like to think in advance about possible obstacles and troubles? Perhaps there are such people, but they are not the ones who create new sequencing methods and do not implement them. nine0003
Who would like to think in advance about possible obstacles and troubles? Perhaps there are such people, but they are not the ones who create new sequencing methods and do not implement them. nine0003
Although in Russia even then there were works that DNA is torn by ultrasound is not accidental. These works were started by Sergei Lvovich Grokhovsky at the Institute of Molecular Biology [10]. It was a very original and original work. Gradually, without NGS sequencing at all, Russian scientists became convinced that DNA was selectively torn by ultrasound [11]. Fortunately, the developers of the NGS methods did not even have time to read the articles of Russian colleagues and widely implemented their technology. And what was available only in one Russian laboratory suddenly appeared in dozens and hundreds of laboratories around the world. The statistics of non-random DNA breakage has become so widespread that the null hypothesis has simply disappeared [12]. And then it turned out that a small methyl group in cytosine makes DNA fragile right next to cytosine [13]. The null hypothesis crackled, on which ideas about the interaction of DNA with regulatory proteins were built. nine0003
The null hypothesis crackled, on which ideas about the interaction of DNA with regulatory proteins were built. nine0003
Something is wrong with cytosine
Cytosine itself makes DNA more fragile, but there is no explanation for this in the usual picture of B-DNA. The sugar-phosphate backbone of DNA breaks, and its structure shows signs of such regularity that one can speak of almost strict periodicity and independence from the nucleotide sequence. The classical double helix provides no clues as to why DNA breaks after cytosine.
Our understanding of DNA began to change quite recently, no more than ten years ago. Short-lived transitional base pairs have been discovered ( transient base pair ) formed in a double helix [14–16]. They turned out to be surprisingly many both in variety and quantity. Some of them, under favorable conditions, could claim more than 1% of the total number of pairs. The observation of such pairs became possible due to the significant development of NMR spectroscopy methods. New methods make it possible to observe states that exist for a few milliseconds, which conventional NMR methods will take for ordinary noise [17]. And the word "observe" is not entirely correct in this case; rather, the method allows you to extract from the noise (dispersion) of the signal some signs of the presence of a particular short-lived pair. It requires a lot of samples, a lot of time and money, and a good model that can explain it all. Yes, we almost forgot: bright heads are still needed. Everything needed was concentrated at Duke University in the laboratory of Professor Al-Hashimi. Within this direction, an attempt has already been made to explain the effect of cytosine methylation - unfortunately, unsuccessful, but very important for us [18]. The fact is that the Al-Hashimi method requires from the very beginning to know what kind of short-lived couple has. We had the opportunity to be the first to understand what this couple should be. nine0003
New methods make it possible to observe states that exist for a few milliseconds, which conventional NMR methods will take for ordinary noise [17]. And the word "observe" is not entirely correct in this case; rather, the method allows you to extract from the noise (dispersion) of the signal some signs of the presence of a particular short-lived pair. It requires a lot of samples, a lot of time and money, and a good model that can explain it all. Yes, we almost forgot: bright heads are still needed. Everything needed was concentrated at Duke University in the laboratory of Professor Al-Hashimi. Within this direction, an attempt has already been made to explain the effect of cytosine methylation - unfortunately, unsuccessful, but very important for us [18]. The fact is that the Al-Hashimi method requires from the very beginning to know what kind of short-lived couple has. We had the opportunity to be the first to understand what this couple should be. nine0003
We used the very idea that there can be non-canonical base pairs in DNA, that is, cytosine can form a non-Watson-Crick pair with guanine. We were looking for a pair that would distort the sugar-phosphate backbone in the vicinity of cytosine and help break this backbone with ultrasound. We had one clue beforehand: we knew exactly what was wrong with cytosine. Cytosine could afford to transform into something similar to uracil. We knew this from the analysis of the NMR spectrum of deoxycytidine: this spectrum contained unremovable signals that could only be explained by the presence of the tautomeric imino form of cytosine [19] (Fig. 3). And this was a dangerous heresy that undermined some dogmas.
We were looking for a pair that would distort the sugar-phosphate backbone in the vicinity of cytosine and help break this backbone with ultrasound. We had one clue beforehand: we knew exactly what was wrong with cytosine. Cytosine could afford to transform into something similar to uracil. We knew this from the analysis of the NMR spectrum of deoxycytidine: this spectrum contained unremovable signals that could only be explained by the presence of the tautomeric imino form of cytosine [19] (Fig. 3). And this was a dangerous heresy that undermined some dogmas.
Tautomers are a common phenomenon in organic chemistry, tautomerism is passed even at school. Most often, tautomerism is just a jumping over of a hydrogen atom inside a molecule, forming a new bond and breaking the old one. But tautomerism in DNA is taboo. If tautomerism is allowed there, then how to explain the accuracy of storage and copying of genetic information? Therefore, the founding fathers acted wisely: if tautomerism is forbidden, then maybe it will not appear? Thus, the irresponsible behavior of the hydrogen atom and the ease of breaking old bonds and forming new ones were condemned. Although we do not approve of such actions as the constant breaking of old and the formation of new bonds in people, but why was it necessary to transfer this condemnation to hydrogen? Some atoms form several bonds at the same time - and can they? However, hydrogen also has hydrogen bonds, so to speak, on the side. Apparently, hydrogen in DNA was condemned just because of them - they wanted the fragile hydrogen bond to be preserved. nine0003
Although we do not approve of such actions as the constant breaking of old and the formation of new bonds in people, but why was it necessary to transfer this condemnation to hydrogen? Some atoms form several bonds at the same time - and can they? However, hydrogen also has hydrogen bonds, so to speak, on the side. Apparently, hydrogen in DNA was condemned just because of them - they wanted the fragile hydrogen bond to be preserved. nine0003
Figure 3. Signs of tautomers. Small bumps in the NMR spectrum of a solution of deoxycytidine (on the vertical axis about 150 and 170) cannot be removed at any pH. It turns out that there are, as it were, five different nitrogen atoms in the cytosine molecule. This can be explained by proton jumping and a change in the nearest environment of two nitrogens at once in the imino tautomer.
Long ago, at the dawn of the era of molecular biology, there were scientists whose prophecies almost always came true.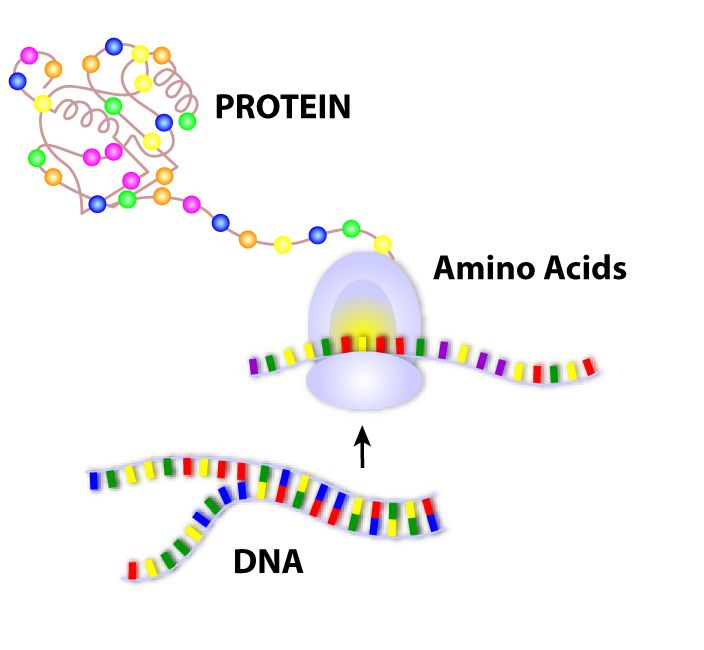 And since the respect for the founding fathers is great, the criticism of the prophecies can be perceived as a criticism of the very demigods who, in molecular biology, are “our everything”. The specific tautomeric form in which base formulas should be written has become a cornerstone in the design of the DNA double helix. The correct entry was suggested to Watson and Crick by Jerry Donoghue [20], who subsequently actively fought against the "dogmas" of molecular biology (including when he was wrong). After the triumph of the DNA double helix, further events can be described as follows:0067 And Francis Crick saw different tautomers and divided them into good ones, capable of entering a double helix, and bad ones, capable of only introducing mutations into it. And Crick commanded that bad tautomers should be rare, and he saw that it was good .
And since the respect for the founding fathers is great, the criticism of the prophecies can be perceived as a criticism of the very demigods who, in molecular biology, are “our everything”. The specific tautomeric form in which base formulas should be written has become a cornerstone in the design of the DNA double helix. The correct entry was suggested to Watson and Crick by Jerry Donoghue [20], who subsequently actively fought against the "dogmas" of molecular biology (including when he was wrong). After the triumph of the DNA double helix, further events can be described as follows:0067 And Francis Crick saw different tautomers and divided them into good ones, capable of entering a double helix, and bad ones, capable of only introducing mutations into it. And Crick commanded that bad tautomers should be rare, and he saw that it was good .
This tautomeric "heaven" and "hell" was probably even a useful assumption at first. Unfortunately, later this began to interfere with the development of understanding, like any dogma.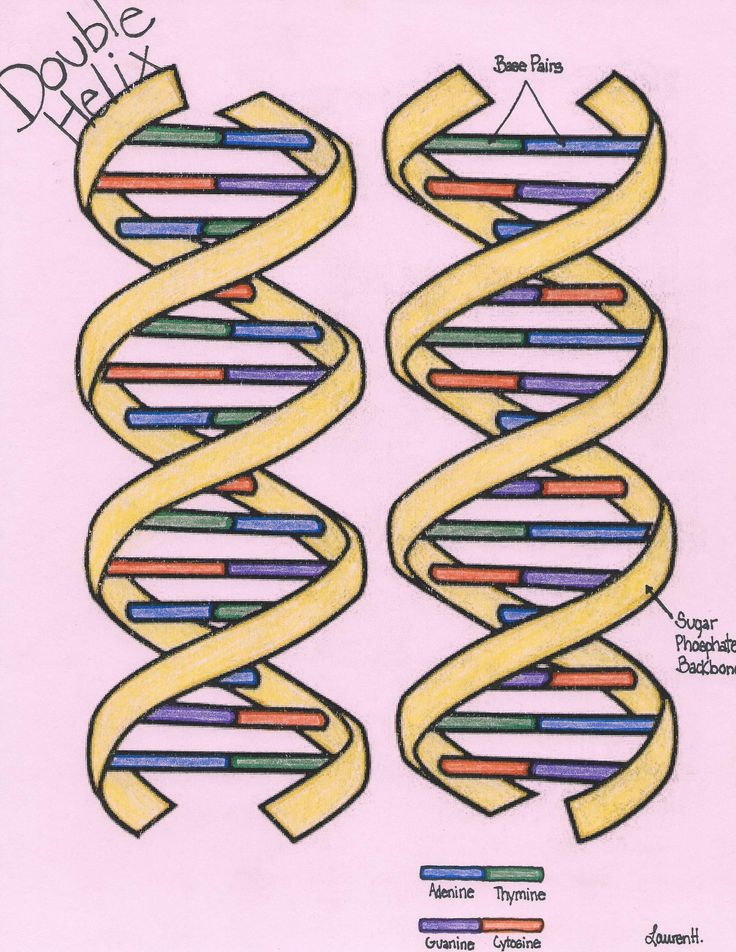 For example, in order not to see tautomers in the NMR spectrum of deoxycytidine (Fig. 3), you can erase their signals from the spectrum, but this would be cheating. In order not to argue with dogma, you can do otherwise: it is enough to spend less time recording the spectrum. There will be more noise, unnecessary signals will be drowned in noise, and reviewers will miss your work. Why record the spectrum for two days, if as a result your manuscript will be returned without review? You can do what our co-author Ilya Eltsov did only if the reviewer in the journal writes to you that he will skip the article when you prove the presence of tautomers in your spectrum. In this work, Ilya was greatly helped by the quarantine announced in the spring, and as a result, a lot of free time appeared on his NMR spectrometer. Ilya was able to take spectra at various pH values, and tautomers were visible everywhere. We express our gratitude to everyone who made quarantine and this work possible. nine0003
For example, in order not to see tautomers in the NMR spectrum of deoxycytidine (Fig. 3), you can erase their signals from the spectrum, but this would be cheating. In order not to argue with dogma, you can do otherwise: it is enough to spend less time recording the spectrum. There will be more noise, unnecessary signals will be drowned in noise, and reviewers will miss your work. Why record the spectrum for two days, if as a result your manuscript will be returned without review? You can do what our co-author Ilya Eltsov did only if the reviewer in the journal writes to you that he will skip the article when you prove the presence of tautomers in your spectrum. In this work, Ilya was greatly helped by the quarantine announced in the spring, and as a result, a lot of free time appeared on his NMR spectrometer. Ilya was able to take spectra at various pH values, and tautomers were visible everywhere. We express our gratitude to everyone who made quarantine and this work possible. nine0003
Talking about the prevalence, importance and even more usefulness of non-canonical pairs involving tautomers was considered bad form until the last decade. Coincidentally, experimental evidence for the prevalence of tautomers began to appear almost simultaneously with data on the prevalence of non-canonical pairs in DNA. It has become not only possible, but also fashionable to write about non-canonical pairs and tautomers, so it is not surprising that articles began to appear where there are non-canonical pairs involving tautomers [7]. It seems that we were just lucky here, but who knows - maybe these are the consequences of some general trend towards combating discrimination? Tautomers and non-canonical couples have ceased to be embarrassed in the last ten years. And it was only a matter of time before these objects showed any use. Tolerance! nine0003
Coincidentally, experimental evidence for the prevalence of tautomers began to appear almost simultaneously with data on the prevalence of non-canonical pairs in DNA. It has become not only possible, but also fashionable to write about non-canonical pairs and tautomers, so it is not surprising that articles began to appear where there are non-canonical pairs involving tautomers [7]. It seems that we were just lucky here, but who knows - maybe these are the consequences of some general trend towards combating discrimination? Tautomers and non-canonical couples have ceased to be embarrassed in the last ten years. And it was only a matter of time before these objects showed any use. Tolerance! nine0003
We took tautomeric cytosine and tried to find a non-canonical pair capable of explaining all the experimental facts, and... found [21].
A pair of wobble-GC: a hybrid of a centaur and a unicorn
The only pair that explained everything observed, turned out to be so incredible that it made me remember two classic English stories at once.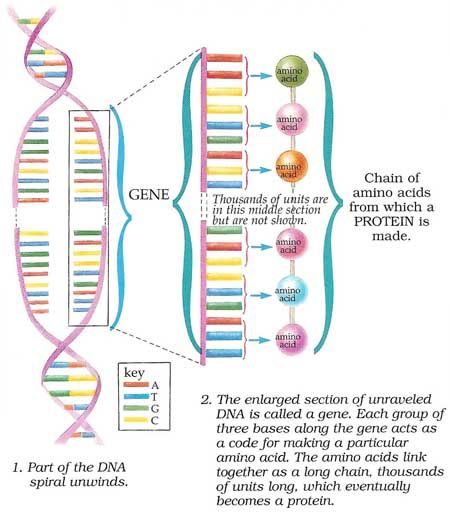 The first is the battle between the Unicorn and the Lion, witnessed by Lewis Carroll's Alice.
The first is the battle between the Unicorn and the Lion, witnessed by Lewis Carroll's Alice.
In order to see the appearance of this pair, look at Figure 4. Does it really resemble our title picture? And it is no coincidence: the sugar atoms C1' are at the same distance - they are, as it were, nailed to the stick with carnations, like figures on a wooden toy. As a result, the displacement of guanine and cytosine into a wobble pair requires that the sugar-phosphate backbone be tensed next to cytosine, but it relaxes (relaxes) near guanine. Cytosine itself has to “tighten up”, since ordinary cytosine does not have the necessary proton for the wobble pair near nitrogen, so it has to turn into a tautomer. nine0003
Note that we have Russified the term wobble here, simply writing it in Cyrillic. The fact is that in Russian there were several modest attempts to translate wobble into the “language of native aspens”: they wrote about “swinging couples”, and about “inaccurate”, even about “ambiguous” - it’s good that dancing couples didn’t offer , swingers and upstart couples (after all, they jump out of the DNA double helix).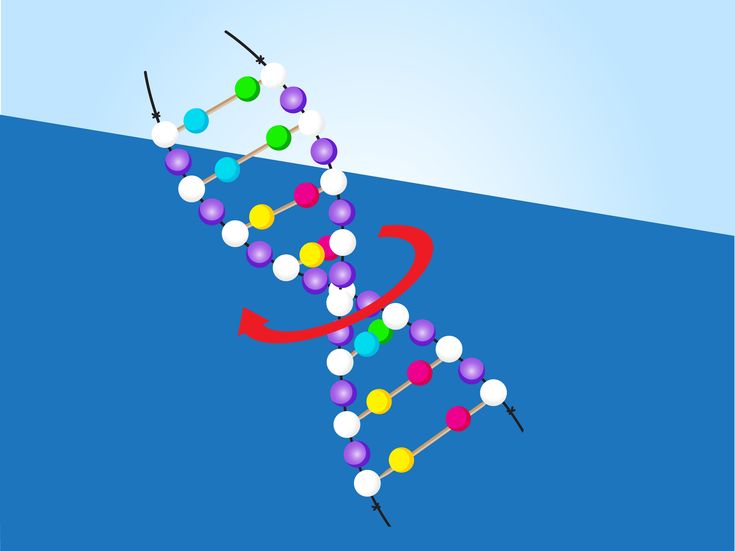 Here we follow Pushkin's testament: " But pantaloons, tailcoat, vest, All these words are not in Russian ". How not? - you say. After all, there are these words in modern Russian! Yes, there are - for Pushkin they appeared recently, like a tracing paper from French. So we are making a simple tracing paper from English, offering a new “clothing” for DNA - such, you know, an epigenetic vest that DNA puts on when it goes to an appointment with epigeneticists.
Here we follow Pushkin's testament: " But pantaloons, tailcoat, vest, All these words are not in Russian ". How not? - you say. After all, there are these words in modern Russian! Yes, there are - for Pushkin they appeared recently, like a tracing paper from French. So we are making a simple tracing paper from English, offering a new “clothing” for DNA - such, you know, an epigenetic vest that DNA puts on when it goes to an appointment with epigeneticists.
Figure 4. Animation depicting the formation of the GC wobble pair
The pair we proposed combined the properties of a tautomer with a wobble structure (Fig. 5). Cytosine becomes an analogue of uracil and forms an analogue of the GU pair. A chimera that combines mythical evil rare tautomers or the legendary wobble pair, sung by Crick in his hypothesis of the same name. Ten years ago, any editor would have pointed out to us that we are mocking everything that is most sacred, because if this is possible in DNA, then any accident is possible? nine0003
It helped us a little that a couple of GUs with tautomers and an exact copy of the Watson-Crick structure have been known for several years [17]. We could say that the couple is short-lived, and this disgrace is not for long. But to hide the fact that this outrage is useful, we could not. There is something contradictory, paradoxical and even absurd in the fact that the wobble structure invented by Francis Crick helps the tautomer rejected by Crick to become an important part of DNA. Of course, this paradox is connected with the undoubted fact that Francis Crick was at the very epicenter of the Big Bang that gave birth to the Universe of molecular biology. Still, we must not forget that Crick was a genius, which means that even his delusions can be perceived as truths for a long time. nine0003
We could say that the couple is short-lived, and this disgrace is not for long. But to hide the fact that this outrage is useful, we could not. There is something contradictory, paradoxical and even absurd in the fact that the wobble structure invented by Francis Crick helps the tautomer rejected by Crick to become an important part of DNA. Of course, this paradox is connected with the undoubted fact that Francis Crick was at the very epicenter of the Big Bang that gave birth to the Universe of molecular biology. Still, we must not forget that Crick was a genius, which means that even his delusions can be perceived as truths for a long time. nine0003
Figure 5. Tautomeric (a) and protonated (b) form of the wobble-GC pair. Normal cytosine does not have a proton to form the damaging hydrogen bond above. But a proton can jump from the amino group (this is a tautomer), or a proton can appear when the medium is acidified (then a charge will appear on nitrogen). I just want to say: "Well, hello, monster!"
The second plot is the classic Sherlock Holmes saying: " If all versions but one are discarded, then this last one, improbable as it may seem, will be the true .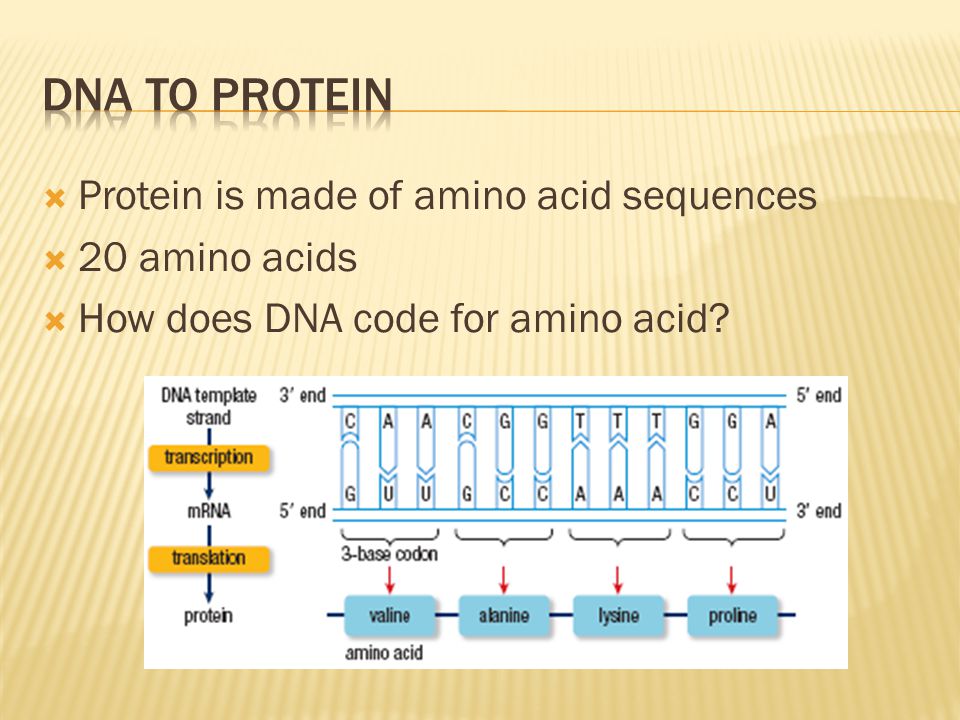 ” Holmes made a logical error here: the last version could also be incorrect and would indicate the inconsistency of the entire chain of reasoning. But we had the opportunity to test the latest version for strength.
” Holmes made a logical error here: the last version could also be incorrect and would indicate the inconsistency of the entire chain of reasoning. But we had the opportunity to test the latest version for strength.
First, we needed to find a non-canonical GC pair that has no analogues based on the AT pair. Secondly, we needed the vapor to significantly and at the same time asymmetrically distort the sugar-phosphate backbone. Thirdly, we needed the possibility of a pair appearing as a result of cytosine protonation. The third condition was caused by the fact that DNA cleavage is greatly enhanced by a moderate decrease in pH [9]. Under these conditions, cytosine is protonated, and protonated cytosine, similarly to tautomeric cytosine, is able to make a wobble pair (Fig. 4). The Hoogsteen pair GC also satisfied the last two conditions, although the wobble pair fulfilled the second condition better. We were lucky: Al-Hashimi relied on the Hoogsteen pair and did not find its connection with methylation [18].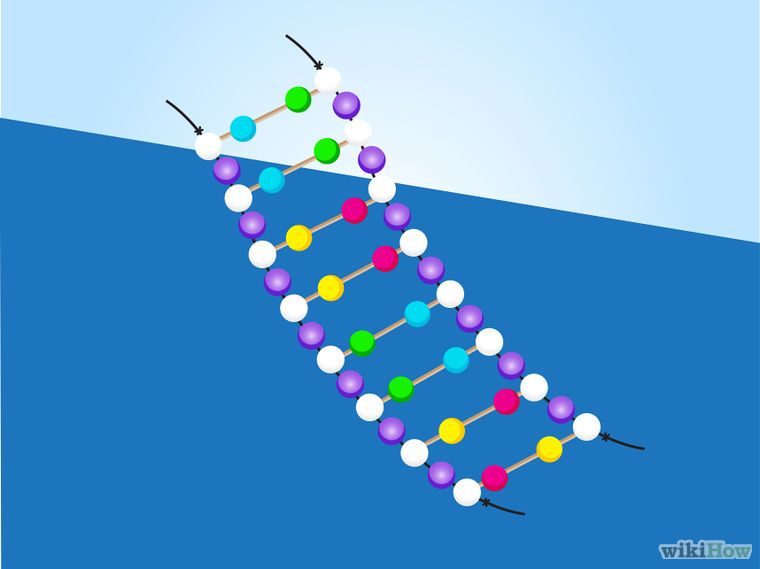
Methylation to control tautomerism
Here we come to the answer to the question: how does cytosine differ from 5-methylcytosine. 5-methylcytosine is more prone to iminotautomer formation. The methyl group as an electron density donor shifts the tautomeric equilibrium towards the formation of a "rare tautomer". This shift provokes the formation of a GC wobble pair (Fig. 4, left), which is reflected in an increase in the frequency of DNA cleavage by ultrasound. A pleasant addition to this idea is the ability to explain the properties of all four modified cytosines, which are suspected or found to be epigenetic markers, by tautomerism [21]. In addition to 5-methylcytosine, we have explained the properties of 5-hydroxymethylcytosine, 5-formylcytosine, and N4-aminomethylcytosine (Fig. 6). nine0003
Figure 6. All cytosine derivatives suspected of complicity in the regulation of gene expression tend to form an iminotautomeric form. ( A ) 5-hydroxymethylcytosine, ( B ) 5-formylcytosine and ( B ) N4-aminomethylcytosine.
Exactly from the same positions it is possible to explain the difference between thymine and uracil. The presence of a methyl group in thymine shifts the equilibrium towards that tautomer, where there is a proton near nitrogen N3, that is, towards the usual keto form of thymine. Why is this needed? Recall that the possibility of forming GU pairs with the participation of a rare tautomer has already been proven, which means that methylation increases the accuracy of DNA copying. But other than that, methylation raises the melting point of the double helix, which can be even more beneficial. nine0003
Well, that's all, we blurted out how thymine differs from uracil. They did not keep the intrigue until the end of the story. Is it interesting to read a detective story after the killer and motive become known? But we've got another page of action, chases and petty fights in store. And also an indication of what to expect in the next season.
Proof from the dark side
Although we found something useful in tautomers and non-canonical pairs, this does not mean that we magically pulled them over to the side of good.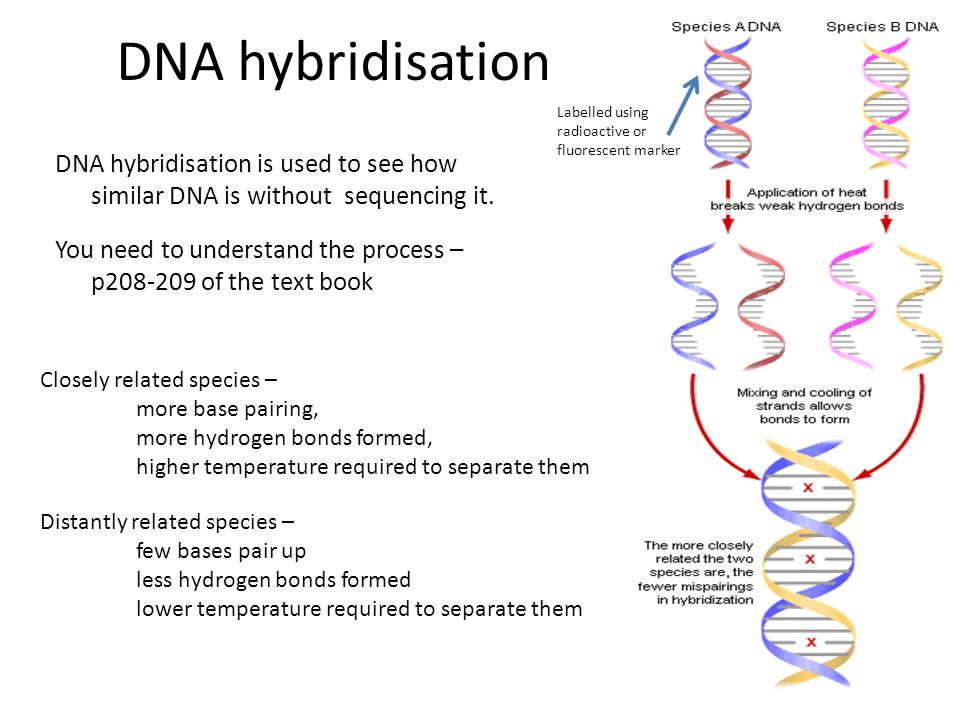 Not only did we not cancel the increased risk of mutations for these pairs, but we were even able to substantiate them anew. The GC of the wobble pair leaves the N4 nitrogen without a hydrogen bond, which means it is completely defenseless against outside attacks (please take a look at Figure 4 again). This nitrogen position increases the likelihood of cytosine deamination. nine0003
Not only did we not cancel the increased risk of mutations for these pairs, but we were even able to substantiate them anew. The GC of the wobble pair leaves the N4 nitrogen without a hydrogen bond, which means it is completely defenseless against outside attacks (please take a look at Figure 4 again). This nitrogen position increases the likelihood of cytosine deamination. nine0003
This can be considered the dark side of tautomerism, but it is more rational to consider mutations as an acceptable price for the ability to regulate, switch and control the DNA molecule. On the one hand, it's a shame that DNA tends to deteriorate just in such an important place. On the other hand, we are not surprised when the part of the mechanism that moves more than the rest breaks down.
Methylation does increase the risk of cytosine deamination mutation. Textbooks and non-fiction literature have established the opinion that the repair system does not cope well with a pair of GTs, as it is used to a pair of GUs.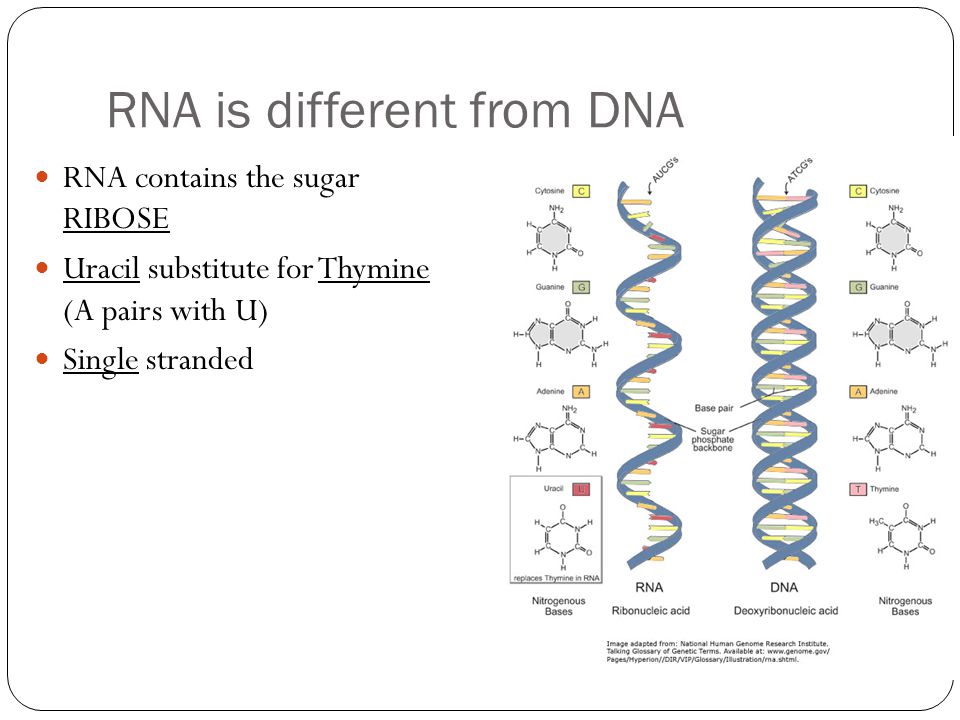 However, there are direct experimental data that indicate an increase in the rate of mutation. And the reparation system, having met GT, must unambiguously determine what needs to be removed T, that is, the problem is solvable. nine0003
However, there are direct experimental data that indicate an increase in the rate of mutation. And the reparation system, having met GT, must unambiguously determine what needs to be removed T, that is, the problem is solvable. nine0003
What does this prove? In the Hoogsteen pair GC, the amino group is stabilized by hydrogen bonding with guanine. A competitor would provoke a completely different mutation. This is another proof that between a wobble and a Hoogsteen pair, we must choose a wobble.
Benefits of the epigenetic edifice from shifting the foundation
Have we damaged the slender epigenetic edifice? Can everything that has already been created by overwork and exorbitant costs of money and reagents for sequencing continue to exist? Is it possible to quickly repair cracks in the walls after shifting the foundation? To be honest, we are not very concerned about the answers to these questions. We look at the current situation in a philistine, pragmatic way: we would like to build an outbuilding or a turret on a shifted foundation. That is, we would like to discuss possible prospects for building epigenetics on a new foundation. For example, the effect of pH on cytosine methylation has not been discussed before, and the mechanism we have discovered makes the effect of pH not only possible, but inevitable. Before our work, there were no ideas about how the very mechanism of DNA regulation through methylation arose, and now it is clear that there were wobble pairs before methylation. Methylation only strengthened, somewhat hypertrophied the existing trend, which means that the mechanism did not appear out of nowhere. The interaction of regulatory proteins with DNA was considered as the recognition of Watson-Crick pairs, and now we have to take into account the jumping out of nucleotides with the possibility of forming new hydrogen bonds. An unexpected prediction was the possible similarity of methyltransferases with repair enzymes, since in both cases it will be necessary to look for a pair in the DNA of a wobble. We feel ourselves, if only for a short time, as those whose prophecies can come true.
That is, we would like to discuss possible prospects for building epigenetics on a new foundation. For example, the effect of pH on cytosine methylation has not been discussed before, and the mechanism we have discovered makes the effect of pH not only possible, but inevitable. Before our work, there were no ideas about how the very mechanism of DNA regulation through methylation arose, and now it is clear that there were wobble pairs before methylation. Methylation only strengthened, somewhat hypertrophied the existing trend, which means that the mechanism did not appear out of nowhere. The interaction of regulatory proteins with DNA was considered as the recognition of Watson-Crick pairs, and now we have to take into account the jumping out of nucleotides with the possibility of forming new hydrogen bonds. An unexpected prediction was the possible similarity of methyltransferases with repair enzymes, since in both cases it will be necessary to look for a pair in the DNA of a wobble. We feel ourselves, if only for a short time, as those whose prophecies can come true. nine0003
nine0003
We are still in that amazing state when the mechanism we have discovered has the status of a hypothesis, which means that it is the duty of real scientists to try to refute our hypothesis. If the hypothesis is strong enough, then we can build all the stones thrown at us into the building that we will build on this new foundation.
The main benefit of the proposed mechanism is the systematic rejection of teleology in epigenetics. DNA is now methylated not “in order to”, but “because”: this process has lost its purpose and a mechanism has appeared. Descriptiveness and contemplation disappeared, causality appeared. Remember: stucco decoration in the theater ceases to be just decoration and becomes a sound-diffusing surface. Behind beauty comes meaning. There is a new view of the whole field of knowledge, and many can now try to look at epigenetics from a new point of view. nine0003
Instead of conclusion
In conclusion, we are forced to make an important reassuring remark: DNA breaking by ultrasound stops completely at a temperature of 30 degrees Celsius - and since our temperature exceeds this value, the process explained in the text is in no way connected with rumors that ultrasound can cause mutations and intrauterine development disorders.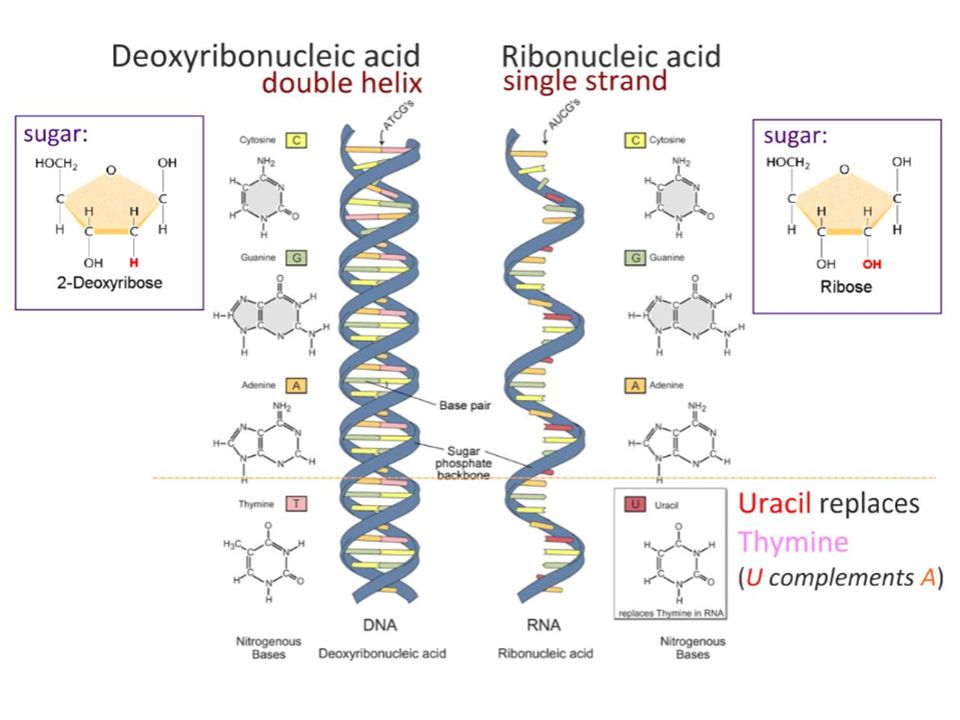 Forgive me if we accidentally interested you in the association with this topic, and now disappointed you.
Forgive me if we accidentally interested you in the association with this topic, and now disappointed you.
The original version of this essay was published in Bioessays [21] .
- How myelin thickness and epigenetics contribute to stress resilience;
- Jail for genes;
- Epigenome pills;
- Epigenetics of psoriasis: molecular marks of fate;
- Cold water, boiled water and boiling milk, or Once again about rejuvenation;
- Aging and longevity: the epigenome reveals mysteries;
- Aging clock: reset, slow down, reverse?; nine0261
- Alberts B., Johnson A., Lewis D. Molecular biology of the cell: with the tasks of Wilson J., Hunt T. Moscow-Izhevsk: Institute for Computer Research, 2013. - 773 p.;
- Sergei L. Grokhovsky, Irina A. Il'icheva, Dmitry Yu. Nechipurenko, Michail V.
 Golovkin, Larisa A. Panchenko, et. al. (2011). Sequence-Specific Ultrasonic Cleavage of DNA. Biophysical Journal . 100 , 117-125;
Golovkin, Larisa A. Panchenko, et. al. (2011). Sequence-Specific Ultrasonic Cleavage of DNA. Biophysical Journal . 100 , 117-125; - Grokhovsky S.L. Specificity of DNA cleavage by ultrasound. Molecular Biology: RAN, 2006. —p. 317-325.; nine0261
- I.A. Ilyichev, Institute of Molecular Biology. V.A. Engelhardt RAS, M.V. Khodykov, L.A. Panchenko, R.V. Polozov, et. al. (2020). ULTRASONIC DNA CLEAVAGE: ANALYSIS OF THE STRUCTURAL AND DYNAMIC CHARACTERISTICS OF THE REGULATORY REGIONS OF THE GENOME AND SEQUENCING ERRORS. Biophysics . 65 , 504-511;
- Maria S. Poptsova, Irina A. Il'icheva, Dmitry Yu. Nechipurenko, Larisa A. Panchenko, Mingian V. Khodikov, et. al. (2015). Non-random DNA fragmentation in next-generation sequencing. nine0029 Sci Rep . 4 ;
- Leonid A. Uroshlev, Eldar T. Abdullaev, Iren R. Umarova, Irina A.
 Il’icheva, Larisa A. Panchenko, et. al. (2020). A Method for Identification of the Methylation Level of CpG Islands From NGS Data. Sci Rep . 10 ;
Il’icheva, Larisa A. Panchenko, et. al. (2020). A Method for Identification of the Methylation Level of CpG Islands From NGS Data. Sci Rep . 10 ; - Garrett B. Goh, Elena N. Laricheva, Charles L. Brooks. (2014). Uncovering pH-Dependent Transient States of Proteins with Buried Ionizable Residues. J. Am. Chem. soc. nine0030 . 136 , 8496-8499;
- Bharathwaj Sathyamoorthy, Honglue Shi, Huiqing Zhou, Yi Xue, Atul Rangadurai, et. al. (2017). Insights into Watson–Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Research . 45 , 5586-5601;
- Isaac J. Kimsey, Katja Petzold, Bharathwaj Sathyamoorthy, Zachary W. Stein, Hashim M. Al-Hashimi. (2015). Visualizing transient Watson–Crick-like mispairs in DNA and RNA duplexes. nine0029 Nature . 519 , 315-320;
- 12 methods in pictures: structural biology;
- Atul Rangadurai, Johannes Kremser, Honglue Shi, Christoph Kreutz, Hashim M.